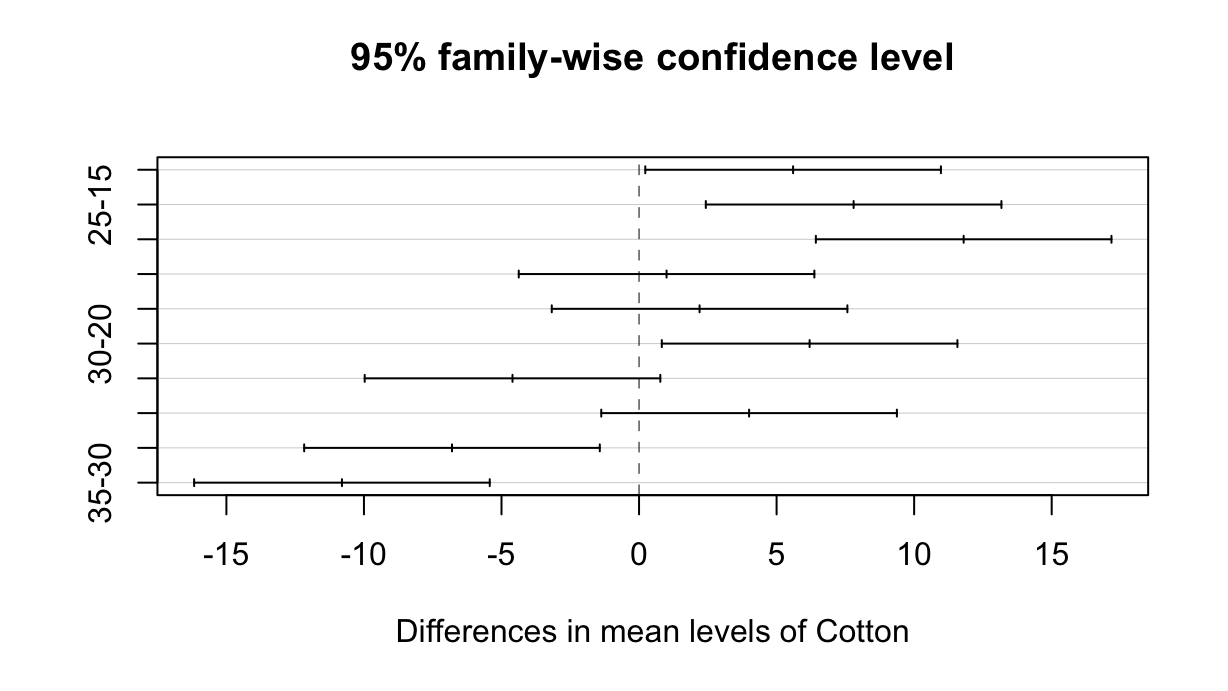
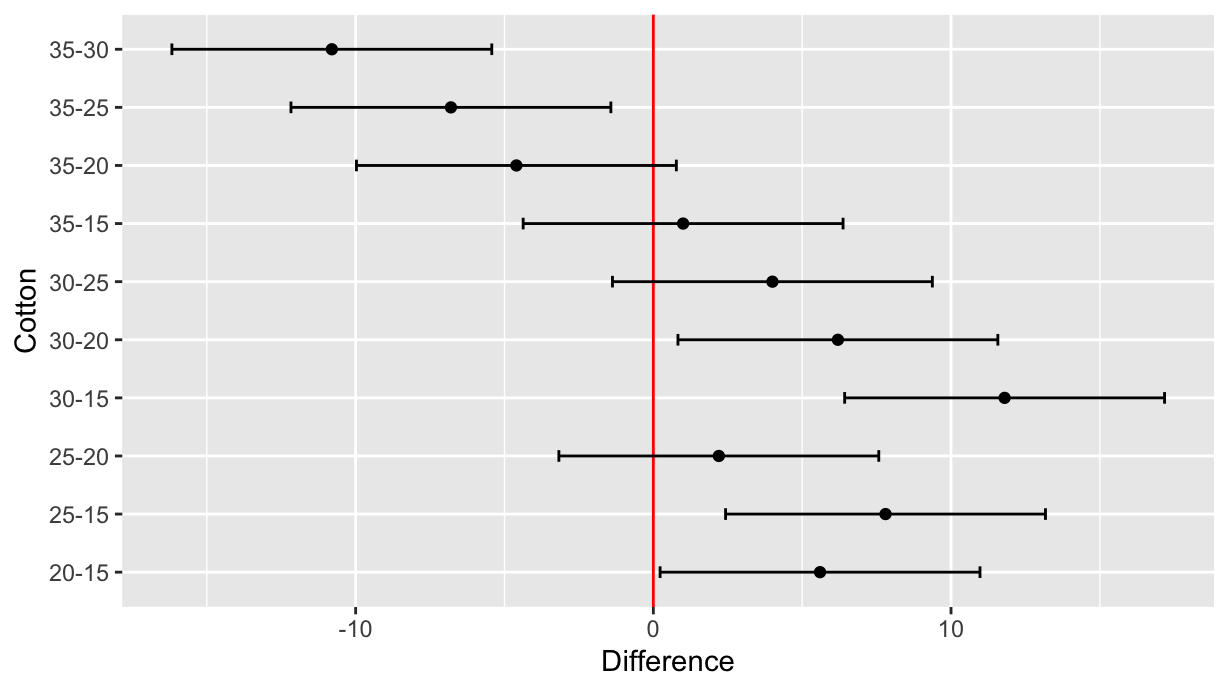
library(tidyverse)
library(adas.utils)
data <- examples_url("anova.dat") %>%
read.table(header=TRUE) %>%
mutate(Cotton=factor(Cotton)) %>%
glimpse()Rows: 25
Columns: 3
$ Cotton <fct> 15, 15, 15, 15, 15, 20, 20, 20, 20, 20, 25, 25, 25, 25, 25…
$ Observation <int> 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5…
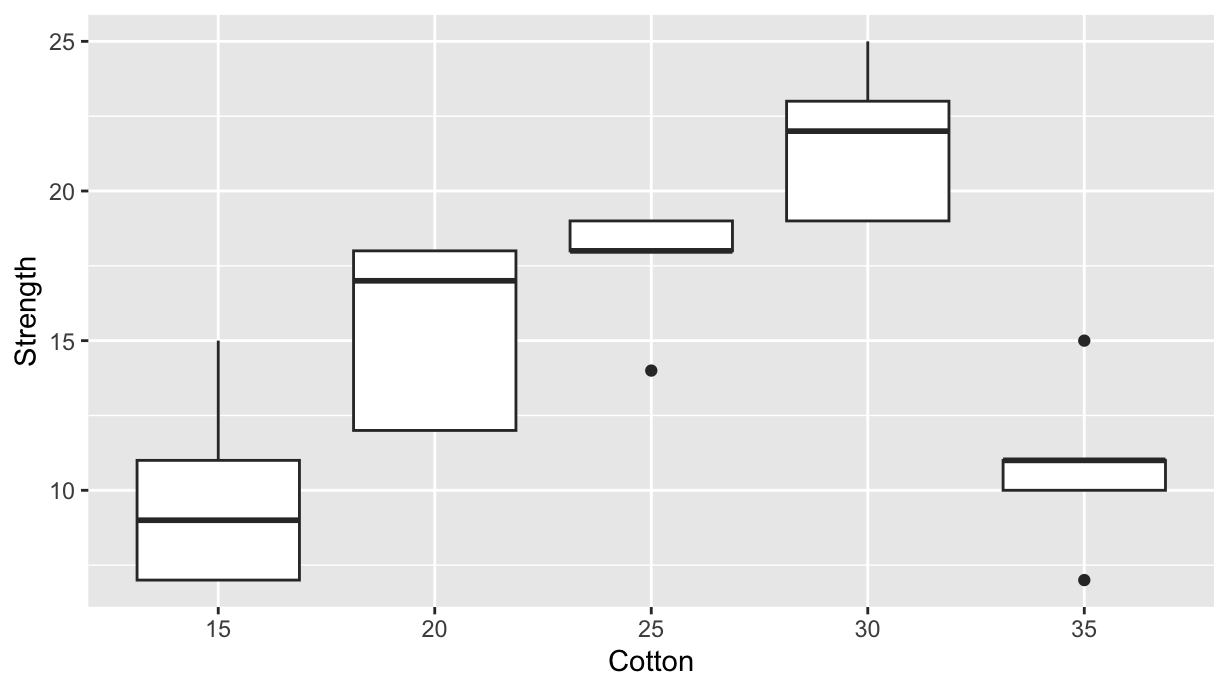
$ Strength <int> 7, 7, 15, 11, 9, 12, 17, 12, 18, 18, 14, 18, 18, 19, 19, 1…data %>%
ggplot(aes(x=Cotton, y=Strength, group=Cotton)) +
geom_boxplot()